Abstract
INTRODUCTION Classic Philadelphia chromosome-negative myeloproliferative neoplasms (MPNs) constitute three phenotypic subtypes - polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF) and are distinguished from other myeloid neoplasms by a combination of morphologic, clinical, laboratory, and cytogenetic/molecular genetic analyses. The existing genetic landscape of MPNs primarily involves mutations in three genes that lead to constitutive JAK-STAT signaling (JAK2, CALR, MPL). Several additional non-driver mutations as well as cytogenetic and epigenetic abnormalities also contribute to disease initiation and progression, and impact both overall survival and potential for progression to acute myeloid leukemia (AML). However, limited understanding exists regarding how genotypic variability contributes to phenotypic presentation and disease natural history (e.g. thrombosis or bleeding; progression from PV or ET to myelofibrosis, or all three diseases to AML). Recent expert review series have called for gene expression profiling, and deeper integration of clinical features, morphology and genetics toward better definition and management of MPNs. Here, we present a proof-of-principle application of transcriptome sequencing technology to characterize disease phenotype in MPN patients, using RNA obtained from disease-relevant peripheral blood cells - platelets.
METHODS MPN patient blood samples were obtained with informed consent and approval by the Stanford Institutional Review Board. To minimize effects of long-term storage of platelets at room temperature and loss of platelet RNA quality and quantity, samples were processed within 6 hours after blood collection. Established methods from the laboratories of Dr. Andrew Weyrich¨ were followed for platelet isolation, purification, and RNA isolation. The integrity of the RNA was evaluated on an Agilent Bioanalyzer, and samples with RNA integrity numbers > 7.0 prepared for sequencing. Sequencing and analysis on platelets from independent human blood samples were performed at the Stanford Functional Genomics facility. Samples were sequenced for 36 cycles, paired-end, on the Illumina GAIIx sequencer. Sequence reads were processed and all downstream analysis performed with a combination of packages available at the Stanford Genomics Bioinformatics Core.
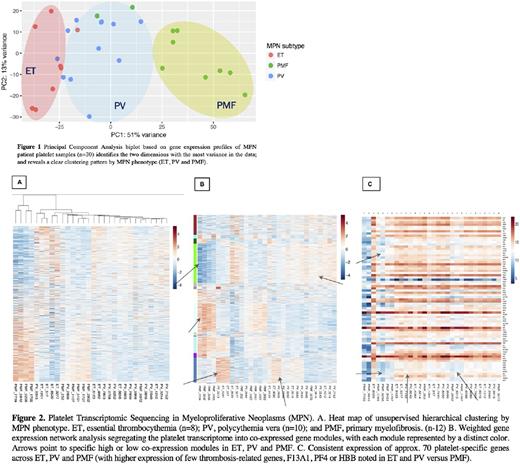
RESULTS Principal component analyses on RNA-sequenced data (Figure 1) reveal clear clustering of gene expression profiles by the three MPN phenotypes - essential thrombocythemia (ET, n=8), polycythemia vera (PV, n=12) and primary myelofibrosis (PMF, n=10). Subsequent bioinformatics analyses included - i) unsupervised hierarchical clustering analysis (Figure 2A), generating a dendrogram that shows distinct segregation of phenotypes on the basisof their gene expression pattern; ii) weighted gene co-expression network analysis (Figure 2B), segregating the platelet transcriptome into co-expressed gene modules [with each module represented by a distinct color; Gene module (green) with high expression in ET (and PV) matched gene ontology terms of "wound healing (GO: 0042060)", "hemostasis (GO:000075999", "blood coagulation (GO: 00007596), and "platelet activation (GO:0030168)"; and interestingly, reflected low expression in PMF patients. On the other hand, GO terms associated with overexpressed genes in PMF patients included "nucleic acid binding (GO:0003676)", "polypyrimidine tract binding (GO: 0008187), "secretory granule organization (GO: 0033363), "axon cargo transport (GO: 0008088)]; and iii) data-mining methods (Figure 2C) to identify 72 relatively platelet-specific/abundant genes including platelet factor-4, platelet basic protein, and coagulation factor 13A1.
CONCLUSIONS We have established a reproducible, high-quality experimental framework for RNA-sequencing of platelets from patients with myeloproliferative neoplasms (MPN). Our preliminary findings reveal unique RNA-based signatures that may help differentiate MPN patients by subtype, mutational status or therapy, and potentially offer a clinically actionable panel for clinical laboratory application . Outcomes from this work may also guide therapeutic strategies toward mitigating thrombosis and bleeding risk in MPN patients.
¨ platelet expert and external collaborator/mentor in this work
Gotlib: Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; CTI Biopharma: Membership on an entity's Board of Directors or advisory committees, Research Funding; Pharmacyclics: Research Funding; Gilead: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Pharmacyclics: Research Funding; Promedior: Research Funding; Deciphera: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Blueprint Medicines: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Incyte: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Promedior: Research Funding; Seattle Genetics: Research Funding; Incyte: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Deciphera: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; CTI Biopharma: Membership on an entity's Board of Directors or advisory committees, Research Funding; Seattle Genetics: Research Funding; Blueprint Medicines: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Gilead: Membership on an entity's Board of Directors or advisory committees, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal